Neb double digest
Through this new partnership we are pleased to offer neb double digest comprehensive next generation sequencing solutions. Unsure of what products are available? Confidently detect more with Archer NGS assay solutions for your solid tumor, blood cancer, immune profiling, and genetic disease research.
Learn more. We are excited to announce that all reaction buffers are now BSA-free. Find more details at www. Web pricing is applicable only to orders placed online at www. Notes Based on the stability of the enzyme in the reaction, incubations longer than 1 hr will not result in improved digestion, unless additional enzyme is added. Please refer to Restriction endonuclease survival in a reaction for more information regarding this topic.
Neb double digest
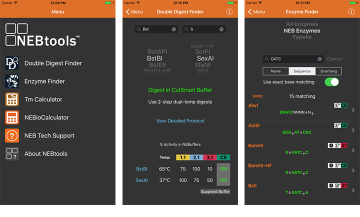
NEBcutter V2. Use this tool to identify the restriction sites within your DNA sequence. Double Digest Finder. Use this tool to guide your reaction buffer selection when setting up double-digests, a common timesaving procedure. Choosing the right buffers will help you to avoid star activity and loss of product. Tm Calculator. Use this tool when designing PCR reaction protocols to help determine the optimal annealing temperature for your amplicon. Simply input your DNA polymerase, primer concentration and your primer sequence and the Tm Calculator will guide you to successful reaction conditions. Options include conversion of mass to moles, ligation amounts, conversion of OD to concentration, dilution and molarity. NEBuilder Assembly Tool. Use this tool to find the right products and protocols for each step digestion, end modification, ligation and transformation of your next traditional cloning experiment. Also, find other relevant tools and resources to enable protocol optimization.
Figure 3. Do you want to sign out? How long the DNA will last depends on its purifty
Ok this is going to be a long post so sit yourself down and prepare to be boggled by this mystery. I am currently trying to insert a 2. The pcDNA 3. I cut out the band using a new razor blade cutting the gel right on the 5. I get a nice clear product only at 5. I then proceed to do a negative control for my vector, so I ligate my vector by itself. After this is done I do a transformation and get tons of colonies in my supposedly negative vector only plate.
Two restriction enzymes are used simultaneously to digest DNA in a single reaction. If your DNA concentration is too low you can increase the reaction volume to ul. Mix well by pipetting slowly up and down approximately 5x. Be gentle, and do not vortex. Spin the samples for 5 seconds in a balanced microcentrifuge, or flick them to collect the mixture at the bottom of the tube. Incubate at 37 degrees for at least 1 hour. For 3A assembly it is important you heat inactivate your samples after digestion. See Section 1.
Neb double digest
We have numerous interactive online tools for these and other questions in your daily lab work. Competitor Cross-Reference Tools. Use this tool to find the right products and protocols for each step digestion, end modification, ligation and transformation of your next traditional cloning experiment.
Brd vs pck dream11 prediction today match
Learn more. If the purpose of the amplification is to add flanking bases to complete a cloning reaction, we strongly recommend ordering the gBlocks HiFi Gene Fragment IDT with the required bases rather than adding them afterwards using PCR. Request a consultation. Use this tool to find the right products and protocols for each step digestion, end modification, ligation and transformation of your next traditional cloning experiment. However, if you require a double-stranded DNA construct outside of this size range, we offer 2 options:. What isoschizomers are there? View FAQ. This rigorous testing ensures that most recombinant colonies obtained from cloning each gBlocks Gene Fragment will contain the desired insert. All gBlocks gene fragments FAQs. Recheck the concentration of your solution. Working on protein or enzyme engineering? Ok this is going to be a long post so sit yourself down and prepare to be boggled by this mystery. If your sequence is not a coding region, we might be able to synthesize the sequence as a custom gene , which is delivered in a basic pUC vector, rather than just a linear, dsDNA fragment.
Web pricing is applicable only to orders placed online at www. Most of our enzymes are supplied with one of four standard NEBuffers.
Application notes and whitepapers. Shipped on dry ice within 10 business days or order confirmation excluding Fridays. Notes Based on the stability of the enzyme in the reaction, incubations longer than 1 hr will not result in improved digestion, unless additional enzyme is added. During PCR, smaller products are more efficiently amplified than longer ones, and at the exponential phase of PCR, smaller products in the reaction will overwhelm the generation of the full-length correct product. If you are using an enzyme that is not supplied with rCutSmart the Performance Chart for Restriction Enzymes rates the percentage activity of each restriction endonuclease in the four standard NEBuffers. While dry, they should be stable up to two years. NEB sells two neoschizomers of KasI. Tm Calculator Use this tool when designing PCR reaction protocols to help determine the optimal annealing temperature for your amplicon. Use this tool to find the right products and protocols for each step digestion, end modification, ligation and transformation of your next traditional cloning experiment. Incorporating 6 NNK codons corresponds to about 1 billion possible combinations, and 18 N mixed bases will create a pool with After resuspending in a high-quality, molecular-grade water or buffer, pH 7. Actually it is 4 small lanes that I combined into one big lane with tape on my comb, 1 lane generally can take 20 microliters Do you think that it is too conc.?

Bravo, brilliant phrase and is duly